Non-alcoholic fatty liver disease, a chronic liver condition not caused by alcohol or other obvious liver-damaging factors, is characterized by excessive fat deposition in hepatocytes. There is a 25% global incidence rate of NAFLD, with those with diabetes and obesity having the highest rates. Various potentially harmful factors significantly influence the development of this chronic illness. NAFLD risk factors include obesity, insulin resistance, and other elements of the metabolic syndrome. Eating patterns, cardiovascular conditions, genetic polymorphisms of numerous genes, etc. are pathogenic factors for NAFLD. NAFLD's exact pathophysiology is yet uncertain, and NAFLD-specific chemotherapy treatments are not yet approved. The FDA does not yet approve treatments for NAFLD. Investigational pharmacotherapies mainly focus on resistance to insulin, the regulation of bile acid in the liver, and lipid management. By employing advanced methods of analysis to identify biomarkers, such as hepatokines, that precisely indicate the effects of exercise training on the functioning of the liver, which may be feasible to monitor the effect of exercise on NAFLD and enhance the commitment of these patients to exercise training. NAFLD is heritable, and multiple loci have been linked to the disease at different stages. Regardless of the fact that implementing a healthy diet and exercising regularly has been found to be beneficial, there are currently no FDA-approved medications for the treatment of NAFLD, and for the majority of patients, making and maintaining these changes has been difficult. The hunt for effective therapeutic medications is a field of research that is currently active. Due to their essential role in the transcriptional regulation of lipid and glucose metabolism, PPAR ligands have been investigated as potential NAFLD therapeutic agents.
The nuclear receptor proteins known as peroxisome proliferator-activated receptors (PPARs) work as transcription factors to control gene expression. PPAR's functions include regulating the genes responsible for fatty acid absorption and oxidation, metabolism of lipids and carbohydrates, inflammation, and proliferation of cells. It also serves as an efficient therapeutic target for the management of numerous forms of metabolic syndrome, particularly NAFLD, diabetes mellitus, and dyslipidemia. PPARα, PPARγ, and PPARβ/δ are three isoforms. Overexpression of PPAR causes NAFLD.
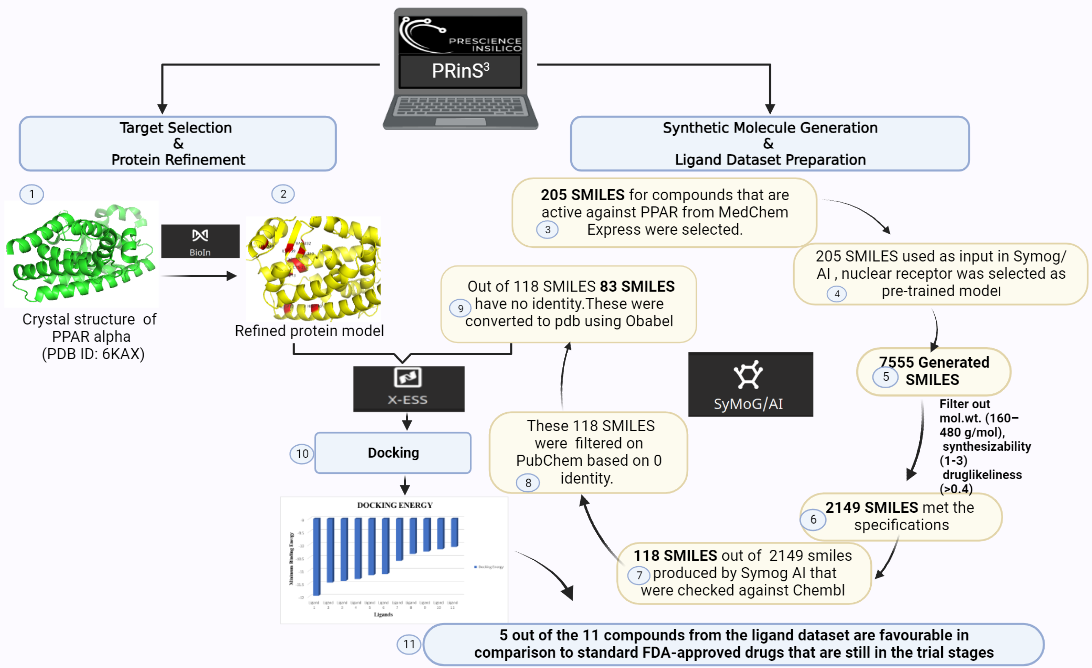
The target protein for this study is Peroxisome proliferator-activated receptor alpha (PPARα), it has a resolution of 1.23Å, the PDB ID 6KAX, and the active site residues as: M330, V332, I339, L247, C275, C276, F273, I447, H440, Y314 was refined using BioIn tool of PRinS3. 205 SMILES for compounds that are active against PPAR were searched using MedChem Express. These 205 Canonical SMILES in .smi format were uploaded, and the pre-trained model was chosen as Nuclear Receptor in the Symog/AI interface. The number of Smiles to be generated was set to 100 , 7555 SMILES were generated. These 7555 SMILES were then filtered on the basis of molecular weight (160–480 g/mol), synthesizability (1-3), and druglikeliness (>0.4). Of the total generated SMILES, 2149 SMILES met the specifications. Of the 2149 smiles produced by Symog AI that were checked against Chembl to identify novel SMILES, 118 were discovered to be novel. A total of 118 SMILES were obtained and filtered based on their PubChem identities. 83 SMILES had no identity. Then, Open Babel software was used to convert each of the 83 SMILES to a PDB file which was then used as a ligand dataset. Using the X-ESS tool, the target protein was docked with the ligand dataset. FDA-approved drugs still in the trial phase were docked with the target protein. To find out the novel molecule against the target protein, first, the compounds were filtered based on the hydrogen bond and minimum binding energy in order to compare them with docked drugs that are still in the trial stages. It has been found that 5 of the 11 compounds in the ligand dataset are better than FDA-approved drugs that are still in the trial stage and can be used for research purposes.
Authors: Megha Bose