Unlocking the Potential of AlphaFold in Drug Discovery: A PRinS3 Perspective
AlphaFold and PRinS3: Transforming Drug Discovery
DeepMind and the EMBL-European Bioinformatics Institute (EMBL-EBI) have formed an exclusive partnership to transform the prediction of three-dimensional (3D) protein structures. This collaboration led to the public release of AlphaFold, a groundbreaking achievement in protein structure prediction. Developed by DeepMind, a Google subsidiary, AlphaFold demonstrated exceptional accuracy in the 14th Critical Assessment of Protein Structure Prediction (CASP14), earning recognition as a breakthrough of the year by Nature (Science’s 2021 Breakthrough of the Year).
The release of over 200 million protein structures by AlphaFold's remarkable performance is reshaping the field of structural biology, with significant implications for biology and medicine. Drug discovery is a critical area that benefits from this advancement, as precise protein structure information is needed to identify drug targets. Despite AlphaFold's major success, researchers have raised concerns about its predicted structures' accuracy for drug discovery.
PRinS3 and BioIn: Enhancing Drug Discovery
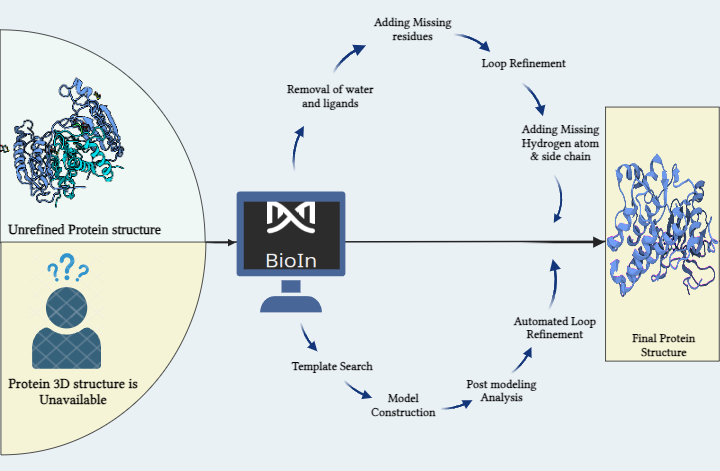
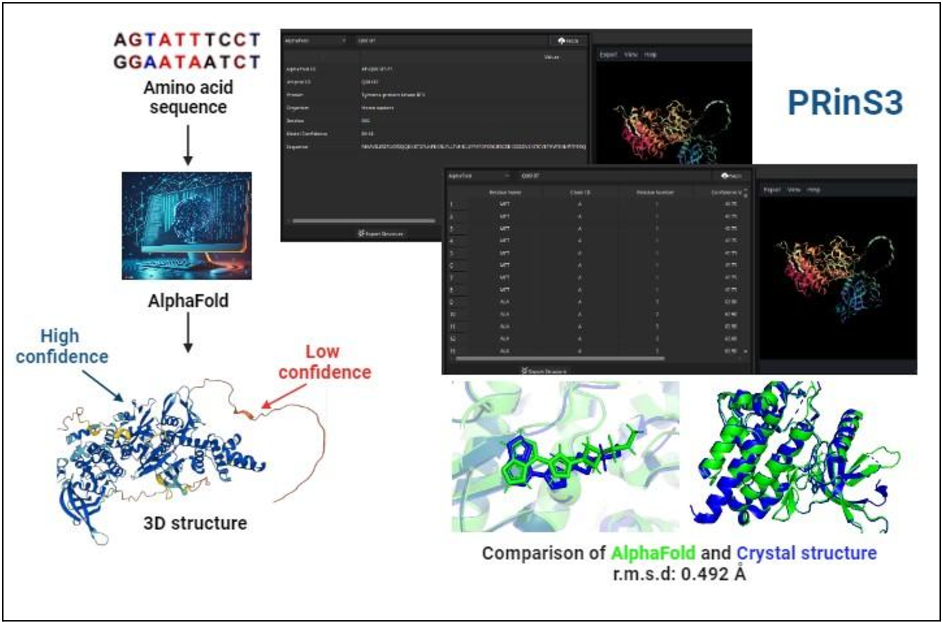
PRinS3, our in-house software suite, is a high-throughput platform designed to host various applications for drug discovery. The BioIn application, a powerful tool specializing in protein refinement and structure prediction, has recently incorporated the AlphaFold database. The database integrated into BioIn offers a range of powerful features:
- Access to Over 200 Million Structures: It provides access to more than 200 million predicted protein structures, covering nearly all known proteins from various species.
- Detailed Protein Information: It includes essential details such as protein names and sequences, making it easy for researchers to find relevant structures.
- Confidence Score: Each predicted protein structure is accompanied by a confidence score (predicted local distance difference test, or pLDDT), ensuring users can assess the reliability of the predicted structures.
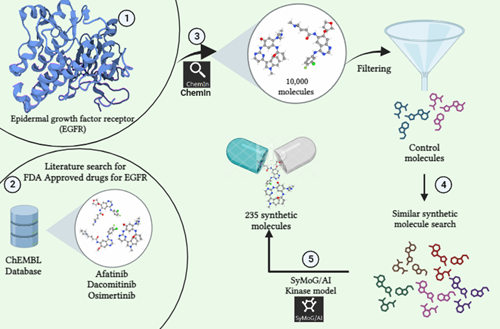
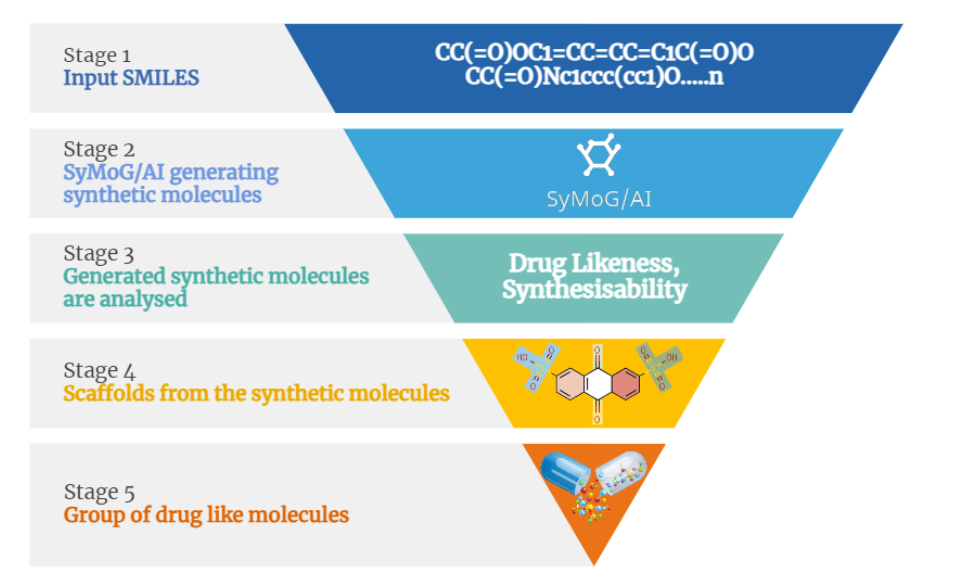
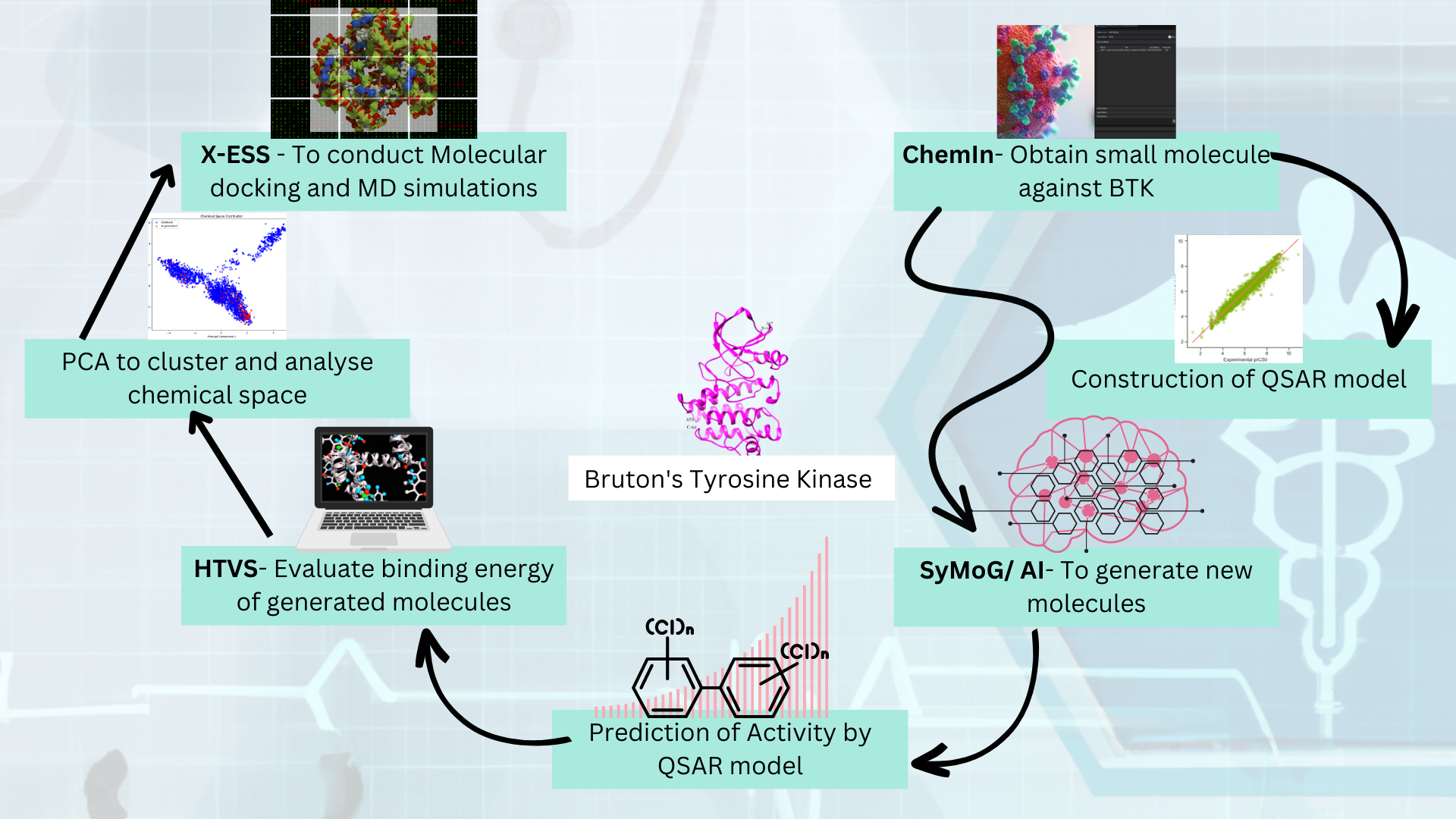
Integrating AlphaFold, alongside access to the RCSB Protein Data Bank (PDB), provides researchers with a comprehensive and powerful set of resources, significantly enhancing the drug discovery process by enabling faster and more accurate identification of potential drug targets. To address this, a comparison was conducted between 102 PDB crystal structures and AlphaFold's predicted structures of kinase proteins. BioIn can be used to refine existing kinase crystal structures by retaining only the necessary chains, removing unwanted chains and water molecules, and keeping only the ligands intact. Similarly, kinase AlphaFold structures relevant to the study were sourced from the AlphaFold Protein Structure Database. The kinase ligand datasets were prepared using publicly available small molecule (ligand) databases.
X-ESS is an automated pipeline designed for high-throughput screening of multiple targets against thousands or even millions of compounds. Capable of processing large datasets, it can simultaneously handle around 500 combinations of targets and ligands. X-ESS conducts docking-based screening, with or without specific binding site information, to deliver valuable insights into target-ligand interactions. This platform significantly accelerates target-based drug development's optimization, screening, and evaluation phases. This study used X-ESS for docking studies of kinase crystal structures and AlphaFold models against various ligands, providing crucial insights into binding energy, scores, and detailed target-ligand interactions.
Our analysis showed that while crystal structures demonstrated slightly higher overall accuracy and binding pocket precision than AlphaFold predictions, discrepancies in ligand poses within AlphaFold structures led to misalignment. Despite AlphaFold's impressive performance in protein structure prediction, these differences suggest that the predicted structures may still need to be more reliable for drug discovery applications without further refinement. Post-processing modifications could improve the quality and consistency of AlphaFold structures, making them more suitable as drug discovery targets.
In conclusion, PRinS3's integrated suite of applications, including BioIn and X-ESS, enabled a comprehensive evaluation of AlphaFold predicted structures for drug discovery. While AlphaFold has revolutionized the field of protein structure prediction, additional research is needed to determine the suitability of its predictions as drug targets. Ongoing efforts to improve the accuracy and consistency of AlphaFold structures, combined with the powerful capabilities of PRinS3, hold great promise for accelerating drug discovery.
With the integration of AI-driven technologies, the landscape of drug discovery is experiencing unprecedented advancements, promising a brighter future. For detailed insights into these cutting-edge applications, we encourage you to explore our official website at https://prescience.in/. To access the platform and obtain essential information for an enhanced experience, please contact us via email at support@prescience.in. Additionally, you can find valuable information on our applications at https://www.prescience.in/prins3/.
Author: Anuradha